Dried Brown Smooth Shiitake Mushroom Dry Shitake,Soaking Dried Shiitake Mushrooms,Rehydrating Shiitake,Dried Sliced Shiitake Mushrooms SHANDONG JOIN & SHARE AGRICULTURAL DEVELOPMENT CO., LTD , https://www.joinsharemushroom.com
1. The overall idea of ​​DNA-based protein detection system 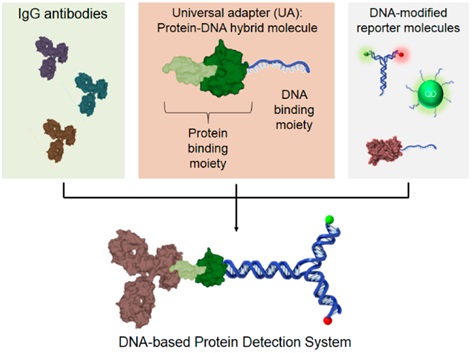
3. Functional verification of UA---binding IgG and DNA-modified labeling molecules 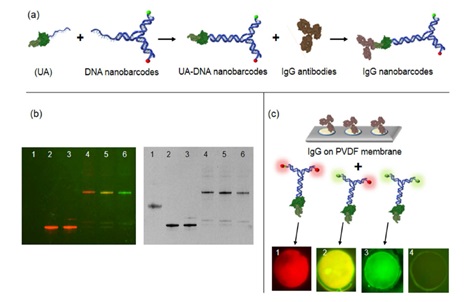
A Universal DNA-Based Protein Detection System
A universal DNA-based protein detection system
Abstract: Antibody-based protein detection methods, such as western blot, ELISA, dot hybridization and immunohistochemistry, are widely used in research and diagnostic fields. In the immunoassay of proteins, the sample protein is first bound to a specific primary antibody, and then detected by a secondary antibody carrying a tag (such as a fluorescent dye, a radioactive element, an enzyme, etc.). However, in order to avoid cross-reactions within the species, the secondary antibody must be carefully selected. At the same time, limited, available primary and secondary antibody pairs further limit the immunoassay of proteins. This article mainly introduces a DNA-based protein detection system that does not require a secondary antibody. The system mainly uses a universal aptamer that binds to DNA-modified reporter molecules and binds to IgG antibodies instead of markers. The secondary antibody allows multiple detections of a single experiment and modularizes the function. DNA-modified Nanobarcodes, quantum dots (QDs), and horseradishperoxidase (HRP) were used as examples to illustrate the DNA-based universal protein detection platform by point hybridization, western blot, immunostaining and microspheres.
Figure 1. (Middle) UA, a bifunctional protein−DNA hybrid molecule, which binds to most types of IgG antibodies (left) and any DNA modified reporter molecules (right) to generate a modular library of pre-labeled primary IgG antibodies for any applications Of protein detection in place of secondary antibodies.
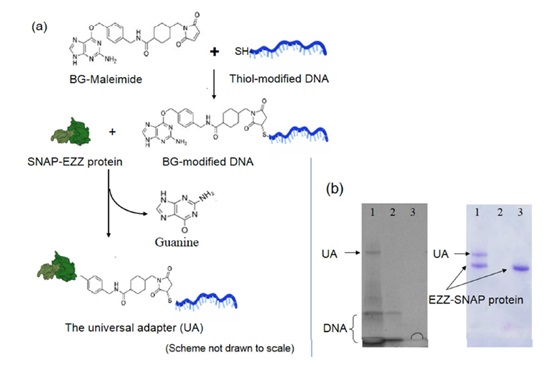
2. Synthesis and identification of UA
(a) Scheme of forming the universal adapter (UA) using a self-catalyzing enzyme (SNAP). Thiol-modified DNA tag was first conjugated to maleimide-benzyl guanine (BG) that served as the substrate for the SNAP enzyme. This BG -modified DNA tag was then linked to EZZ protein through SNAP catalysis to create the universal adapter (UA), a bifunctional protein-DNA hybrid molecule. (b) Confirmation of UA on 4-20% SDS-PAGE gel electrophoresis. The same gel Was first stained with GelRed dye (left) to visualize DNA products and then stained with Comassie Brilliant Blue (right) to visualize protein products. Only UA with both DNA and protein components is visualized in both cases. Lane 1. UA; Lane 2. Thiol-modified DNA only; 3. EZZ-SNAP protein only.
(a) Scheme to confirm the binding of UA against DNA nanobarcodes and IgG primary antibodies. DNA linker of UA was hybridized with DNA nanobarcodes to form UA-DNA nanobarcodes. UA-DNA nanobarcodes bound to IgG antibodies to form IgG nanobarcodes. Characterizing of the hybridization of UA and DNA nanobarcodes on native PAGE gel electrophoresis. The same gel with no staining (left) showed signals of distinct color ratios from DNA nanobarcodes, and with GelRed nucleic acid stain (right) showed signals from all DNA products. Lane 1. UA; Lane 2. R2G0-DNA nanobarcode; Lane 3. EZZ protein mixed with R2G0-DNA nanobarcode; Lanes 4-6. UA hybridized with R2G0-, R1G1-, and R0G2-DNA nanobarcodes, respectively. In lanes 4-6 showed that DNA nanobarcodes were linked to UA through DNA hybridization. (c) Characterizations of the binding of UA-DNA nanobarcodes to IgG antibodies using dot blot technique. Different IgG antibodies were spotted on PVDF membrane and incubated with the UA - DNA nanobarcodes. Dots 1-3. Rabbit polyclonal antibodies (anti-glucagon, anti-PDX-1, and anti-insulin) labelled with R2G0-, R1G1-, and R0G2-nanobarcodes, respectively. Fluorescence signals detected from dots 1-3 Showing strong binding between the IgG antibodies and EZZ protein of UA. Dot 4. Goat polyclonal antibody (anti-PDX-1) labelled with R1G1-nanobarcode. When using a non-specific IgG that cannot bind with EZZ protein, there was no fluorescence Signal detected from dot 4. Dot diameters were 250 Km for all images.
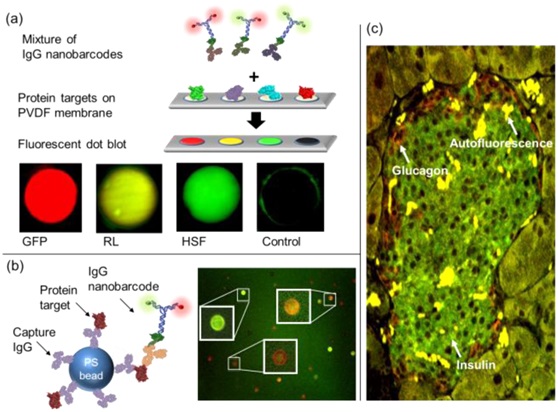
4. Protein detection---Detecting the corresponding proteins by dot hybridization, microspheres and immunostaining using IgG nanobarcodes
Proteins were spotted on a PVDF membrane and then incubated with a detection solution, which contained a mixture of IgG nano-barcodes. The dots with IgG nano-barcodes for protein detection. GFP, RL, HSF, and sortase (negative control) proteins were labeled with R2G1-anti-RL, and R1G1-anti-RL, and R0G2-anti-HSF IgG antibodies, respectively. Dot diameters were 250 μm for all images. Background fluorescence from GFP alone was subtracted from the measurement with IgG nano-barcodes. (b) Multiplexed bead-based protein detection. PS beads were tagged with The first IgG antibody which captured the antigen and then sandwiched with IgG nano-barcodes. GFP, RL, and HSF protein targets were specifically detected on microbeads with R2G0-anti-GFP (red), R1G1-anti-RL IgG antibodies (orange) , and R0G2-anti-HSF ( (c) IgG nanobarcodes for immunostaining of insulin and glucagon proteins in a diabetic mouse pancreas tissue. Insulin was stained with R0G2-antiinsulin (green) antibody, and glucagon was stained with R2G0-antiglucagon (red) antibody. Intense Yellow regions are the autofluorescence of contaminated red blood cells.
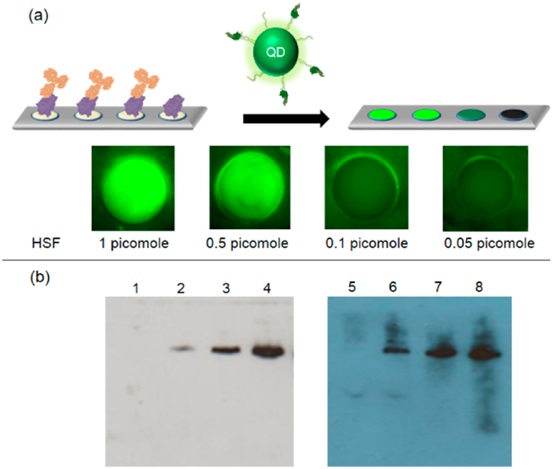
5. Protein detection---Detecting HSF protein by dot blot and western blot using IgG-UA-QDs and IgG-UA-HRP
Figure 3. DNA-based protein detection system with different reporter molecules. (a) Protein detection using dot blot technique with quantum dots. Quantum dots were used to replace DNA nanobarcodes in our IgG nano-barcodes. HSF protein on PVDF membrane was recognized by Its specific antibody and then labeled with UA-QDs. (b) Comparison of WB detection of HSF protein using DNA-modified HRP with IgG-UA (IgG-UA-HRP) (left) and traditional HRPmodified secondary antibody (right). 1−5, HSF protein detection using our IgG-UA-HRP at 0.25, 0.5, 0.75, 1, and 2 pmol; lanes 6−9, HSF protein detection using traditional WB method which utilized the HRP-modified secondary antibody at 0.25, 0.5, 0.75, and 1pmol.